Overview of the SAMPL6 pKa challenge: evaluating small molecule microscopic and macroscopic pKa predictions
/Mehtap Işık, Ariën S Rustenburg, Andrea Rizzi, Marilyn R Gunner, David L Mobley, John D Chodera
Journal of Computer-Aided Molecular Design 35:131, 2021
[DOI] [bioRxiv] [GitHub] [manuscript and figure sources]
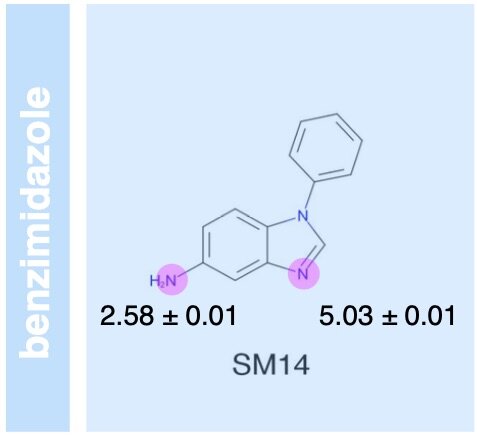
The SAMPL6 pKa challenge assessed the ability of the computational chemistry community to predict macroscopic and microscopic pKas for a set of druglike molecules resembling kinase inhibitors. This paper reports on the overall performance and lessons learned, including the surprising finding that many tools predict reasonably accurate macroscopic pKas corresponding to the wrong microscopic protonation sites.